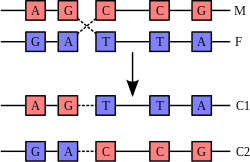
In genetics, complementary DNA (cDNA) is DNA synthesized from a mature mRNA template in a reaction catalysed by the enzyme reverse transcriptase. cDNA is often used to clone eukaryotic genes in prokaryotes.
Overview
The central dogma of molecular biology outlines that in synthesizing proteins, DNA is transcribed into mRNA, which is translated into protein. One difference between eukaryotic and prokaryotic genes is that eukaryotic genes can contain introns (intervening sequences), which are not coding sequences, and must be spliced out of the RNA primary transcript before it becomes mRNA and can be translated into protein. Prokaryotic genes have no introns, so their RNA is not subject to splicing.
Often it is desirable to express eukaryotic genes in prokaryotic cells. A simplified method of doing so would include the addition of eukaryotic DNA to a prokaryotic host, which would transcribe the DNA to mRNA and then translate it to protein. However, as eukaryotic DNA has introns, and since prokaryotes lack the machinery to splice them, the splicing of eukaryotic DNA must be done prior to adding the eukaryotic DNA into the host. This spliced DNA is called complementary DNA. To obtain expression of the protein encoded by the eukaryotic cDNA, prokaryotic regulatory sequences would also be required (e.g. a promoter).
Synthesis
Though there are several methods for doing so, cDNA is most often synthesized from mature (fully spliced) mRNA using the enzyme reverse transcriptase. This enzyme operates on a single strand of mRNA, generating its complementary DNA based on the pairing of RNA base pairs (A, U, G and C) to their DNA complements (T, A, C and G respectively).
To obtain eukaryotic cDNA whose introns have been spliced:
- A eukaryotic cell transcribes the DNA (from genes) into RNA (pre-mRNA).
- The same cell processes the pre-mRNA strands by splicing out introns, and adding a poly-A tail and 5’ Methyl-Guanine cap.
- This mixture of mature mRNA strands are extracted from the cell.
- A poly-T oligonucleotide primer is hybridized onto the poly-A tail of the mature mRNA template, or random hexamer primers can be added which contain every possible 6 base single strand of DNA and can therefore hybridize anywhere on the RNA (Reverse transcriptase requires this double-stranded segment as a primer to start its operation.)
- Reverse transcriptase is added, along with deoxynucleotide triphosphates (A, T, G, C).
The reverse transcriptase scans the mature mRNA and synthesizes a sequence of DNA that complements the mRNA template. This strand of DNA is complementary DNA.
Note that the central dogma of molecular biology is not broken in this process.
Applications
Complementary DNA is often used in gene cloning or as gene probes or in the creation of a cDNA library. Partial sequences of cDNAs are often obtained as expressed sequence tags.
Viruses
Some viruses also use cDNA to turn their viral RNA into mRNA (viral RNA → cDNA → mRNA). The mRNA is used to make viral proteins to take over the host cell.
http://en.wikipedia.org/wiki/Complementary_DNA




![The DNA structure at left (schematic shown) will self-assemble into the structure visualized by atomic force microscopy at right. DNA nanotechnology is the field which seeks to design nanoscale structures using the molecular recognition properties of DNA molecules. Image from Strong, 2004. [1]](http://upload.wikimedia.org/wikipedia/commons/thumb/5/55/DNA_nanostructures.png/400px-DNA_nanostructures.png)