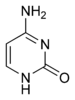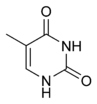
The chemical structure of DNA.
DNA is a long polymer made from repeating units called nucleotides.[1][2] The DNA chain is 22 to 26 Ångströms wide (2.2 to 2.6 nanometres), and one nucleotide unit is 3.3 Ångstroms (0.33 nanometres) long.[3] Although each individual repeating unit is very small, DNA polymers can be enormous molecules containing millions of nucleotides. For instance, the largest human chromosome, chromosome number 1, is 220 million base pairs long.[4]
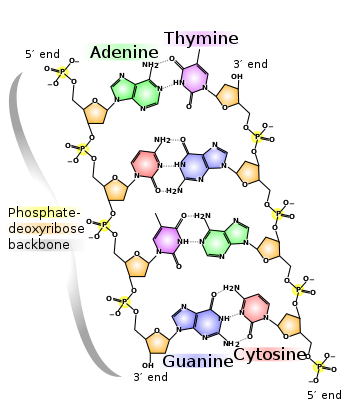
In living organisms, DNA does not usually exist as a single molecule, but instead as a tightly-associated pair of molecules.[5][6] These two long strands entwine like vines, in the shape of a double helix. The nucleotide repeats contain both the segment of the backbone of the molecule, which holds the chain together, and a base, which interacts with the other DNA strand in the helix. In general, a base linked to a sugar is called a nucleoside and a base linked to a sugar and one or more phosphate groups is called a nucleotide. If multiple nucleotides are linked together, as in DNA, this polymer is referred to as a polynucleotide.[7]
The backbone of the DNA strand is made from alternating phosphate and sugar residues.[8] The sugar in DNA is 2-deoxyribose, which is a pentose (five-carbon) sugar. The sugars are joined together by phosphate groups that form phosphodiester bonds between the third and fifth carbon atoms of adjacent sugar rings. These asymmetric bonds mean a strand of DNA has a direction. In a double helix the direction of the nucleotides in one strand is opposite to their direction in the other strand. This arrangement of DNA strands is called antiparallel. The asymmetric ends of DNA strands are referred to as the 5′ (five prime) and 3′ (three prime) ends. One of the major differences between DNA and RNA is the sugar, with 2-deoxyribose being replaced by the alternative pentose sugar ribose in RNA.[6]
The DNA double helix is stabilized by hydrogen bonds between the bases attached to the two strands. The four bases found in DNA are adenine (abbreviated A), cytosine (C), guanine (G) and thymine (T). These four bases are shown below and are attached to the sugar/phosphate to form the complete nucleotide, as shown for adenosine monophosphate.
These bases are classified into two types; adenine and guanine are fused five- and six-membered heterocyclic compounds called purines, while cytosine and thymine are six-membered rings called pyrimidines.[6] A fifth pyrimidine base, called uracil (U), usually takes the place of thymine in RNA and differs from thymine by lacking a methyl group on its ring. Uracil is not usually found in DNA, occurring only as a breakdown product of cytosine, but a very rare exception to this rule is a bacterial virus called PBS1 that contains uracil in its DNA.[9] In contrast, following synthesis of certain RNA molecules, a significant number of the uracils are converted to thymines by the enzymatic addition of the missing methyl group. This occurs mostly on structural and enzymatic RNAs like transfer RNAs and ribosomal RNA.[10]
Major and minor grooves

Animation of the structure of a section of DNA. The bases lie horizontally between the two spiraling strands.
Large version[11] The double helix is a right-handed spiral. As the DNA strands wind around each other, they leave gaps between each set of phosphate backbones, revealing the sides of the bases inside (see animation). There are two of these grooves twisting around the surface of the double helix: one groove, the major groove, is 22 Å wide and the other, the minor groove, is 12 Å wide.[12] The narrowness of the minor groove means that the edges of the bases are more accessible in the major groove. As a result, proteins like transcription factors that can bind to specific sequences in double-stranded DNA usually make contacts to the sides of the bases exposed in the major groove.[13]
Base pairing
At top, a
GC base pair with three
hydrogen bonds. At the bottom,
AT base pair with two hydrogen bonds. Hydrogen bonds are shown as dashed lines.
Each type of base on one strand forms a bond with just one type of base on the other strand. This is called complementary base pairing. Here, purines form hydrogen bonds to pyrimidines, with A bonding only to T, and C bonding only to G. This arrangement of two nucleotides binding together across the double helix is called a base pair. In a double helix, the two strands are also held together via forces generated by the hydrophobic effect and pi stacking, which are not influenced by the sequence of the DNA.[14] As hydrogen bonds are not covalent, they can be broken and rejoined relatively easily. The two strands of DNA in a double helix can therefore be pulled apart like a zipper, either by a mechanical force or high temperature.[15] As a result of this complementarity, all the information in the double-stranded sequence of a DNA helix is duplicated on each strand, which is vital in DNA replication. Indeed, this reversible and specific interaction between complementary base pairs is critical for all the functions of DNA in living organisms.[1]
The two types of base pairs form different numbers of hydrogen bonds, AT forming two hydrogen bonds, and GC forming three hydrogen bonds (see figures, left). The GC base pair is therefore stronger than the AT base pair. As a result, it is both the percentage of GC base pairs and the overall length of a DNA double helix that determine the strength of the association between the two strands of DNA. Long DNA helices with a high GC content have stronger-interacting strands, while short helices with high AT content have weaker-interacting strands.[16] Parts of the DNA double helix that need to separate easily, such as the TATAAT Pribnow box in bacterial promoters, tend to have sequences with a high AT content, making the strands easier to pull apart.[17] In the laboratory, the strength of this interaction can be measured by finding the temperature required to break the hydrogen bonds, their melting temperature (also called Tm value). When all the base pairs in a DNA double helix melt, the strands separate and exist in solution as two entirely independent molecules. These single-stranded DNA molecules have no single common shape, but some conformations are more stable than others.[18]
Sense and antisense
A DNA sequence is called "sense" if its sequence is the same as that of a messenger RNA copy that is translated into protein. The sequence on the opposite strand is complementary to the sense sequence and is therefore called the "antisense" sequence. Since RNA polymerases work by making a complementary copy of their templates, it is this antisense strand that is the template for producing the sense messenger RNA. Both sense and antisense sequences can exist on different parts of the same strand of DNA (i.e. both strands contain both sense and antisense sequences). In both prokaryotes and eukaryotes, antisense RNA sequences are produced, but the functions of these RNAs are not entirely clear.[19] One proposal is that antisense RNAs are involved in regulating gene expression through RNA-RNA base pairing.[20]
A few DNA sequences in prokaryotes and eukaryotes, and more in plasmids and viruses, blur the distinction made above between sense and antisense strands by having overlapping genes.[21] In these cases, some DNA sequences do double duty, encoding one protein when read 5′ to 3′ along one strand, and a second protein when read in the opposite direction (still 5′ to 3′) along the other strand. In bacteria, this overlap may be involved in the regulation of gene transcription,[22] while in viruses, overlapping genes increase the amount of information that can be encoded within the small viral genome.[23] Another way of reducing genome size is seen in some viruses that contain linear or circular single-stranded DNA as their genetic material.[24][25]
Supercoiling
DNA can be twisted like a rope in a process called DNA supercoiling. With DNA in its "relaxed" state, a strand usually circles the axis of the double helix once every 10.4 base pairs, but if the DNA is twisted the strands become more tightly or more loosely wound.[26] If the DNA is twisted in the direction of the helix, this is positive supercoiling, and the bases are held more tightly together. If they are twisted in the opposite direction, this is negative supercoiling, and the bases come apart more easily. In nature, most DNA has slight negative supercoiling that is introduced by enzymes called topoisomerases.[27] These enzymes are also needed to relieve the twisting stresses introduced into DNA strands during processes such as transcription and DNA replication.[28]

From left to right, the structures of A, B and Z DNA
Alternative double-helical structures
DNA exists in several possible conformations. The conformations so far identified are: A-DNA, B-DNA, C-DNA, D-DNA,[29] E-DNA,[30] H-DNA,[31] L-DNA,[29] P-DNA,[32] and Z-DNA.[8][33] However, only A-DNA, B-DNA, and Z-DNA have been observed in naturally occurring biological systems. Which conformation DNA adopts depends on the sequence of the DNA, the amount and direction of supercoiling, chemical modifications of the bases and also solution conditions, such as the concentration of metal ions and polyamines.[34] Of these three conformations, the "B" form described above is most common under the conditions found in cells.[35] The two alternative double-helical forms of DNA differ in their geometry and dimensions.
The A form is a wider right-handed spiral, with a shallow, wide minor groove and a narrower, deeper major groove. The A form occurs under non-physiological conditions in dehydrated samples of DNA, while in the cell it may be produced in hybrid pairings of DNA and RNA strands, as well as in enzyme-DNA complexes.[36][37] Segments of DNA where the bases have been chemically-modified by methylation may undergo a larger change in conformation and adopt the Z form. Here, the strands turn about the helical axis in a left-handed spiral, the opposite of the more common B form.[38] These unusual structures can be recognized by specific Z-DNA binding proteins and may be involved in the regulation of transcription.[39]

Structure of a DNA quadruplex formed by
telomere repeats. The conformation of the DNA backbone diverges significantly from the typical helical structure
[40]
Quadruplex structures
At the ends of the linear chromosomes are specialized regions of DNA called telomeres. The main function of these regions is to allow the cell to replicate chromosome ends using the enzyme telomerase, as the enzymes that normally replicate DNA cannot copy the extreme 3′ ends of chromosomes.[41] As a result, if a chromosome lacked telomeres it would become shorter each time it was replicated. These specialized chromosome caps also help protect the DNA ends from exonucleases and stop the DNA repair systems in the cell from treating them as damage to be corrected.[42] In human cells, telomeres are usually lengths of single-stranded DNA containing several thousand repeats of a simple TTAGGG sequence.[43]
These guanine-rich sequences may stabilize chromosome ends by forming very unusual structures of stacked sets of four-base units, rather than the usual base pairs found in other DNA molecules. Here, four guanine bases form a flat plate and these flat four-base units then stack on top of each other, to form a stable G-quadruplex structure.[44] These structures are stabilized by hydrogen bonding between the edges of the bases and chelation of a metal ion in the centre of each four-base unit. The structure shown to the left is a top view of the quadruplex formed by a DNA sequence found in human telomere repeats. The single DNA strand forms a loop, with the sets of four bases stacking in a central quadruplex three plates deep. In the space at the centre of the stacked bases are three chelated potassium ions.[45] Other structures can also be formed, with the central set of four bases coming from either a single strand folded around the bases, or several different parallel strands, each contributing one base to the central structure.
In addition to these stacked structures, telomeres also form large loop structures called telomere loops, or T-loops. Here, the single-stranded DNA curls around in a long circle stabilized by telomere-binding proteins.[46] At the very end of the T-loop, the single-stranded telomere DNA is held onto a region of double-stranded DNA by the telomere strand disrupting the double-helical DNA and base pairing to one of the two strands. This triple-stranded structure is called a displacement loop or D-loop.[44]
http://en.wikipedia.org/wiki/DNA