All the functions of DNA depend on interactions with proteins. These protein interactions can be non-specific, or the protein can bind specifically to a single DNA sequence. Enzymes can also bind to DNA and of these, the polymerases that copy the DNA base sequence in transcription and DNA replication are particularly important.
DNA-binding proteins
 |
 |
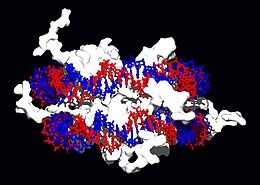
Structural proteins that bind DNA are well-understood examples of non-specific DNA-protein interactions. Within chromosomes, DNA is held in complexes with structural proteins. These proteins organize the DNA into a compact structure called chromatin. In eukaryotes this structure involves DNA binding to a complex of small basic proteins called histones, while in prokaryotes multiple types of proteins are involved.[72][73] The histones form a disk-shaped complex called a nucleosome, which contains two complete turns of double-stranded DNA wrapped around its surface. These non-specific interactions are formed through basic residues in the histones making ionic bonds to the acidic sugar-phosphate backbone of the DNA, and are therefore largely independent of the base sequence.[74] Chemical modifications of these basic amino acid residues include methylation, phosphorylation and acetylation.[75] These chemical changes alter the strength of the interaction between the DNA and the histones, making the DNA more or less accessible to transcription factors and changing the rate of transcription.[76] Other non-specific DNA-binding proteins found in chromatin include the high-mobility group proteins, which bind preferentially to bent or distorted DNA.[77] These proteins are important in bending arrays of nucleosomes and arranging them into more complex chromatin structures.[78]
A distinct group of DNA-binding proteins are the single-stranded-DNA-binding proteins that specifically bind single-stranded DNA. In humans, replication protein A is the best-characterised member of this family and is essential for most processes where the double helix is separated, including DNA replication, recombination and DNA repair.[79] These binding proteins seem to stabilize single-stranded DNA and protect it from forming stem loops or being degraded by nucleases.
In contrast, other proteins have evolved to specifically bind particular DNA sequences. The most intensively studied of these are the various classes of transcription factors, which are proteins that regulate transcription. Each one of these proteins bind to one particular set of DNA sequences and thereby activates or inhibits the transcription of genes with these sequences close to their promoters. The transcription factors do this in two ways. Firstly, they can bind the RNA polymerase responsible for transcription, either directly or through other mediator proteins; this locates the polymerase at the promoter and allows it to begin transcription.[81] Alternatively, transcription factors can bind enzymes that modify the histones at the promoter; this will change the accessibility of the DNA template to the polymerase.[82]
As these DNA targets can occur throughout an organism's genome, changes in the activity of one type of transcription factor can affect thousands of genes.[83] Consequently, these proteins are often the targets of the signal transduction processes that mediate responses to environmental changes or cellular differentiation and development. The specificity of these transcription factors' interactions with DNA come from the proteins making multiple contacts to the edges of the DNA bases, allowing them to "read" the DNA sequence. Most of these base-interactions are made in the major groove, where the bases are most accessible.[84]
DNA-modifying enzymes
Nucleases and ligases
Nucleases are enzymes that cut DNA strands by catalyzing the hydrolysis of the phosphodiester bonds. Nucleases that hydrolyse nucleotides from the ends of DNA strands are called exonucleases, while endonucleases cut within strands. The most frequently-used nucleases in molecular biology are the restriction endonucleases, which cut DNA at specific sequences. For instance, the EcoRV enzyme shown to the left recognizes the 6-base sequence 5′-GAT|ATC-3′ and makes a cut at the vertical line. In nature, these enzymes protect bacteria against phage infection by digesting the phage DNA when it enters the bacterial cell, acting as part of the restriction modification system.[86] In technology, these sequence-specific nucleases are used in molecular cloning and DNA fingerprinting.
Enzymes called DNA ligases can rejoin cut or broken DNA strands, using the energy from either adenosine triphosphate or nicotinamide adenine dinucleotide.[87] Ligases are particularly important in lagging strand DNA replication, as they join together the short segments of DNA produced at the replication fork into a complete copy of the DNA template. They are also used in DNA repair and genetic recombination.[87]
Topoisomerases and helicases
Topoisomerases are enzymes with both nuclease and ligase activity. These proteins change the amount of supercoiling in DNA. Some of these enzyme work by cutting the DNA helix and allowing one section to rotate, thereby reducing its level of supercoiling; the enzyme then seals the DNA break.[27] Other types of these enzymes are capable of cutting one DNA helix and then passing a second strand of DNA through this break, before rejoining the helix.[88] Topoisomerases are required for many processes involving DNA, such as DNA replication and transcription.[28]
Helicases are proteins that are a type of molecular motor. They use the chemical energy in nucleoside triphosphates, predominantly ATP, to break hydrogen bonds between bases and unwind the DNA double helix into single strands.[89] These enzymes are essential for most processes where enzymes need to access the DNA bases.
Polymerases
Polymerases are enzymes that synthesise polynucleotide chains from nucleoside triphosphates. They function by adding nucleotides onto the 3′ hydroxyl group of the previous nucleotide in the DNA strand. As a consequence, all polymerases work in a 5′ to 3′ direction.[90] In the active site of these enzymes, the nucleoside triphosphate substrate base-pairs to a single-stranded polynucleotide template: this allows polymerases to accurately synthesise the complementary strand of this template. Polymerases are classified according to the type of template that they use.
In DNA replication, a DNA-dependent DNA polymerase makes a DNA copy of a DNA sequence. Accuracy is vital in this process, so many of these polymerases have a proofreading activity. Here, the polymerase recognizes the occasional mistakes in the synthesis reaction by the lack of base pairing between the mismatched nucleotides. If a mismatch is detected, a 3′ to 5′ exonuclease activity is activated and the incorrect base removed.[91] In most organisms DNA polymerases function in a large complex called the replisome that contains multiple accessory subunits, such as the DNA clamp or helicases.[92]
RNA-dependent DNA polymerases are a specialised class of polymerases that copy the sequence of an RNA strand into DNA. They include reverse transcriptase, which is a viral enzyme involved in the infection of cells by retroviruses, and telomerase, which is required for the replication of telomeres.[93][41] Telomerase is an unusual polymerase because it contains its own RNA template as part of its structure.[42]
Transcription is carried out by a DNA-dependent RNA polymerase that copies the sequence of a DNA strand into RNA. To begin transcribing a gene, the RNA polymerase binds to a sequence of DNA called a promoter and separates the DNA strands. It then copies the gene sequence into a messenger RNA transcript until it reaches a region of DNA called the terminator, where it halts and detaches from the DNA. As with human DNA-dependent DNA polymerases, RNA polymerase II, the enzyme that transcribes most of the genes in the human genome, operates as part of a large protein complex with multiple regulatory and accessory subunits.[94]
http://en.wikipedia.org/wiki/DNA


No comments:
Post a Comment