Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the double helix winds to the left in a zig-zag pattern (instead of to the right, like the more common B-DNA form). Z-DNA is thought to be one of three biologically active double helical structures along with A- and B-DNA.
History
Z-DNA was the first crystal structure of a DNA molecule to be solved (see: x-ray crystallography). It was solved by Alexander Rich and co-workers in 1979 at MIT.[1] The crystallisation of a B- to Z-DNA junction in 2005[2] provided a better understanding of the potential role Z-DNA plays in cells. Whenever a segment of Z-DNA forms, there must be B-Z junctions at its two ends, interfacing it to the B-form of DNA found in the rest of the genome.
In 2007, the RNA version of Z-DNA was described as a transformed version of an A-RNA double helix into a left-handed helix.[3]
Structure
Z-DNA is quite different from the right-handed forms. In fact, Z-DNA is often compared against B-DNA in order to illustrate the major differences. The Z-DNA helix is left handed and has a structure that repeats every 2 base pairs. The major and minor grooves, unlike A- and B-DNA, show little difference in width. Formation of this structure is generally unfavourable, although certain conditions can promote it; such as alternating purine-pyrimidine sequence, DNA supercoiling or high salt and some cations. Z-DNA can form a junction with B-DNA in a structure which involves the extrusion of a base pair.
Predicting Z-DNA structure
It is possible to predict the likelihood of a DNA sequence forming a Z-DNA structure. An algorithm for predicting the propensity of DNA to flip from the B-form to the Z-form, ZHunt, was written by Dr. P. Shing Ho in 1984 (at MIT). This algorithm was later developed by Tracy Camp, P. Christoph Champ, Sandor Maurice, and Jeffrey M. Vargason for genome-wide mapping of Z-DNA (with P. Shing Ho as the principal investigator).[4] Z-Hunt is available at Z-Hunt online.
Biological significance
While no definitive biological significance of Z-DNA has been found, it is commonly believed to provide torsional strain relief (supercoiling) while DNA transcription occurs.[5][2] The potential to form a Z-DNA structure also correlates with regions of active transcription. A comparison of regions with a high sequence-dependent, predicted propensity to form Z-DNA in human chromosome 22 with a selected set of known gene transcription sites suggests there is a correlation.[4]
Z-DNA formed after transcription initiation in some cases may be bound by RNA modifying enzymes which then alter the sequence of the newly-formed RNA [1].
Comparison Geometries of Some DNA Forms
| Geometry attribute | A-form | B-form | Z-form |
|---|---|---|---|
| Helix sense | right-handed | right-handed | left-handed |
| Repeating unit | 1 bp | 1 bp | 2 bp |
| Rotation/bp | 33.6° | 35.9° | 60°/2 |
| Mean bp/turn | 10.7 | 10.0 | 12 |
| Inclination of bp to axis | +19° | −1.2° | −9° |
| Rise/bp along axis | 2.3 Å (0.23 nm) | 3.32 Å (0.332 nm) | 3.8 Å (0.38 nm) |
| Pitch/turn of helix | 24.6 Å (2.46 nm) | 33.2 Å (3.32 nm) | 45.6 Å (4.56 nm) |
| Mean propeller twist | +18° | +16° | 0° |
| Glycosyl angle | anti | anti | C: anti, G: syn |
| Sugar pucker | C3'-endo | C2'-endo | C: C2'-endo, G: C2'-exo |
| Diameter | 26 Å (2.6 nm) | 20 Å (2.0 nm) | 18 Å (1.8 nm) |
http://en.wikipedia.org/wiki/Z-DNA

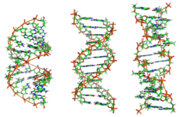

No comments:
Post a Comment